
Citation: Galdino AS, et al. Fungal Phytase Production in Different Hosts: A Brief Review. J Microbiol Biotechnol, 2017, 2(2): 000120.
*Corresponding author: Alexsandro S Galdino, Microbial Biotechnology Laboratory, Universidade Federal de São João Del-Rei, Divinópolis, MG, Brazil 35501-2296, Tel: +55 (37) 3250-5494; Email: asgaldino@ufsj.edu.br
In this mini review we describe the main results of biotechnology studies on fungal phytases available in the literature, their main host cells and mutagenicity methodologies in order to expand our knowledge on fungal phytases produced in different host systems.
Keywords: Phytase; Filamentous Fungi; Heterologous expression
The phytate (phytic acid) present in plants, especially in cereals, is an anti-nutrient that chelates metals and reduces its absorption during digestion in monogastric animals. Excretion of undigested phytate can cause serious ecological problems due to phosphorus excess. Phytases are acid phytic-degrading enzymes used in animal feed supplementation. The great majority of phytases used in animal feed are from fungal origin due to important biochemical properties of these enzymes, such as: thermostability at high temperatures, optimal activity in acidic conditions, and resistance to proteolysis of stomach enzymes such as pepsin and trypsin (Table 1). Taken together, these characteristics in a unique enzyme make these proteins as an important input in industrial animal feed. The main goal of this review article was describe the main hosts used (Figure 1) for expression of recombinant fungal phytases, their advantages and the tools currently used, to generate new phytases with potent industrial properties.
BioreactorsSince Escherichia coli is often used for heterologous expression researchers have dedicated efforts to produce fungal phytase in E. coli in soluble form. This strategy is highly desirable once it can help in high throughput screening of gene libraries constructed by directed evolution. Ushasree, et al. (2014) [1] performed the gene cloning and soluble expression of an Aspergillus niger NII 08121 phytase in E. coli in cytosol via co-expression of chaperones GroES/EL for improving cytosolic solubility of enzymes. This strategy could result in soluble and functional protein products. Alteration in its pH profile indicated the role of glycosylation conserving its characteristic properties [2-5] studying a histidine acid phosphatase (HAP) family phytases (rPhyXT52) from a southern pine beetle fungus garden showed (Dendroctonus frontalis) high enzymatic activity when be expressed in E.coli. Biochemical characterization has shown that phytase is tolerant to high temperatures. When compared to the disulfide bonds, the noncovalent interaction of the salt bridges might play more important roles in the heat-resilient property of these enzymes. The optimum pH (3.9) of the PhyXT52 is close to the usual gastric pH condition of livestock and poultry. E. colias host cells have advantages of easy cloning, maintenance and the formation of inclusion bodies can be bypassed with the co-expressed chaperones. However, the absence of glycosylation remains a disadvantage of the system.
YeastsThe yeasts Pichiapastoris and Saccharomyces cerevisiae have was an interesting alternative as unicellular host cells due to their coatings glycosylated proteins in different patterns. P. pastoris has been used as a system of expression by several researchers Han and Lei (1999), Shi, et al. (2009), Wang, et al. (2007), Zhao DM, et al. (2007) Ullah, et al. (2000), Maldonado, et al. (2014) and Xiong, et al. (2006) [6-12]. They verified the expression of phytase from Aspergillus niger, A. fumigatus, A. japonicas and, Peniophoralycii. The results show that A. niger, A. fumigatus, A. japonicas phytases showed an improvement in their thermostability directly related to glycosylation. This enzyme showed a reduction in molecular mass, thermostability, enzymatic activity and alteration in the optimum pH when was deglycosylated. In contrast, phytase of P. lycii expressed showed no gain in its thermostability even having 10 potential glycosylation sites. The yeast S. cerevisia has been used as an expression system by several researchers worldwide Yanming, et al. (1999) and Wyss, et al. (1999) [13,14]. They verified the phytase expression of A. niger, A. fumigatus and Aspergillus terreus. Wyss, et al. (1999) [14] reported a phytase expressed in S. cerevisia that exhibited excessive glycosylation patterns. However, this excess of glycosylation did not affected the specific activity of the enzyme, the thermostability or the native folding. In another work the A. niger phytase showed a high thermostability when compared to the previous ones due to the high glycosylation range. In this way it is observed that the glycosylation pattern of P. pastoris has improved the thermostability of most of the phytases expressed when compared to phytases expressed in S. cerevisiae.
FungiThe expression system in filamentous fungi has the advantage of high enzymatic production and the various post-transcriptional modifications, but as a disadvantage it has a variable pattern of glycosylation. The fungi A. niger and A. oryzae have been a profitable alternative capable of producing thermostable proteins due to their post-transcriptional machinery. The fungi A. niger expression system has been used expression by several researchers [14-18]. They have verified the expression of phytases from A. ficuum, A. terreus, A. fumigatus, Emericella nidulans, Myceliophthora thermophile and, A. niger. The results showed that the glycosylation patterns were highly variable, differing individually. A high thermostability was reported, for a A. fumigatus phytase, maintaining 90% of enzymatic activity at 100° C for 20 minutes. A. oryzae also has been used as a good expression system by some researchers Uchida, et al. (2006), Ullah and Sethumadhavan (2003) and Lassen, et al. (2001) [19-21]. They checked the phytase expression of A. oryzae, P. lycii, Agrocybe pediades, Ceriporia sp. and Trametes pubescens. The results showed phytases with high thermostability and restricted pH range (4.0-7.0). An important finding was reported by Lassen, et al. (2001) [21-23], basidomycete phytases has preference for attack on phytic acid 6-phosphate, a characteristic never observed in fungi.
Fungi such as A. awamori, H. polymorpha, Penicillium griseoroseum, Kluyveromyces lactis and Fusarium venenatum were used as an expression system others researchers Martin, et al. (2006), Martin, et al. (2003), Wyss, et al. (1999), Corrêa, et al. (2015), Ushasree, et al. (2015), Berka, et al. (1991) [24-29]. They have verified the expression from phytases of A. awamori, A. fumigatus, A. terreus, Talaromyces thermophilus, P. chrysogenum, A. niger, and Thermomyces lanuginosus. Based on this work the homologous expression of a P. chrysogenum phytase expressed in P. griseoroseum, highly stable phytase at room temperature for months.
PlantsThe genus Nicotianahas been studied as an expression system by several researchers Ullah, et al. (1999), George et al. (2005) and Oh et al. (2014) [10,30,31]. They verified the expression of phytase enzymes from A. ficuum, A. niger and A. nidulans. The results have shown the possibility of overexpressing the phyA gene from Aspergillus in other commercial crop plants as an alternative for production of these enzymes [20]. Cloned and expressed the phyA gene in Medicago sativa (alfalfa) leaves. The kinetic parameters of the phyA gene gave nearly identical values to those of the native phytase. Phillippy and Mullaney (1997) [32] verified that phyA gene from when expressed in E. coli was shown to be stored in inclusion bodies and lacked activity. Attempts were made to refold the protein with concomitant regeneration of the activity but without success. This could be due to the lack of glycosylation of fungal phytase after expression in E. coli. Which can be bypassed by the expression system in glycosylating plants. Other types of plants have also been addressed for the expression of heterologous phytase. Plants such as Triticum aestivum, Chlamydomonas reinhardtii, Mature maize, Brassica napus and Zea mays L were used to express A. japonicas phytases reported by Abid, et al. (2017) and A. niger studied by Rao, et al. (2016), Rao J, et al. (2013), Peng, et al. (2006) and Drakakaki, et al. (2005) [33-37] respectively.
Other organism: Silkworm and MicroalgaeThe use of silk worms is an attractive technological alternative for protein expression, once that the pupae are bioreactors of silk production. Xu, et al. (2014) [38] demonstrated the use of transgenic silk worms, Bombyxmori, which was transformed with a codonoptimized A. niger phytase gene (phyA) under the control of the Bmlp3 promoter. The result of this work suggested this system as a potential, "bioreactors" for phyA expression with biomass being produced with low-costs. Microalgae also have high nutritional value. Erpel, et al. (2016) [39] developed a transgenic microalgae (Chlamydomonas reinhardtii) expressing an improved version of the PhyA gene of A. niger, to be used as a food supplement for monogastric animals. This research also tackled the nutritional problems regarding phosphorus deficiency and general animal nutrition.
Mutagenesis ToolsPhytase-directed mutagenesis of A. niger increased the specific activity of phytases in the pH 4-5 changing glutamic acid (E) by lysine (K) at position 300 (K300E) [40]. Also was reported an improvement in their thermostability through the changes T314S, Q315R, V62N clone P9 and S205N, S206A, T151A, T314S, Q315R clone P12 [11]. Changes in the amino acids Q53R and K91D caused an increasing of the enzymatic activity at pH 5.0 and a high affinity to substrate [41]. Changes in the amino acids P212H S238D T255E G377T and D461N caused a change in the interaction of amino acids H82 and Asp362 from the catalytic site, favorably altering the profile of the optimum pH. Conversely, this changes affected negatively the thermostability of the enzyme [28]. Random mutagenesis by error-prone PCR (ep-PCR) in A. niger phytase increased the catalytic efficiency and reduced its thermostability, when the amino acids changes E156G, Q396RT236A and Q396 were made [42] . The site directed mutagenesis of I44E and T252R improved the thermostability and enzyme activity [43]. Random changes in phytase of Penicillium sp. in different clones (T11A, G56E, L65F, Q144H and L151S) and (T11A, H37Y, G56E, L65F, Q144H, L151S and N354D) resulted in an gain of enzyme regarding to thermostability and resistance to pepsin [44], The authors believe that new hydrogen bonds, improved the interaction of the secondary protein structures, reinforcing a possible explanation on protein thermal stability. Hybridization of A. terreus phytase with A. niger showed an increase of phytase thermostability when compared to wild type [16].
Conclusion and PerspectiveThe present review article showed several fungi phytases produced in different hosts as biofactories. In addition, this work has shown the main biochemical properties which are performed in order to obtain innovative products and thus, generate new phytases. One successful strategy is the site directed mutagenesis described previously. Finally, we hope that this article based on fugal phytases can expand our knowledge on recombinant fungal phytases expressed in different hosts
AcknowledgementsThis work was supported by Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG) Grant APQ-00835- 14, PRONEM/FAPEMIG. AS Galdino and NS Parachin thanks to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for its research fellowship.
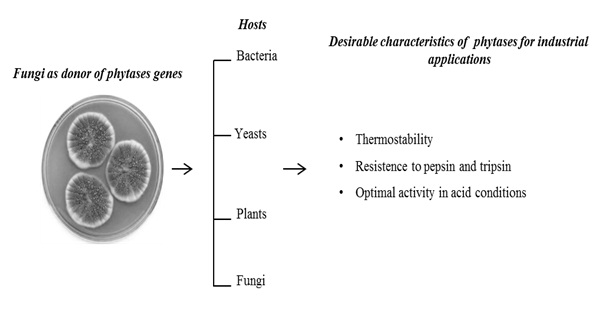
Figure 1: The graph representation of the main hosts used for fungal phytases production and its important biochemical properties for the developing of new phytases for industrial applications. The Penicillium picture was taken from web site https://lovecraftianscience.wordpress.com/tag/penicillium/.
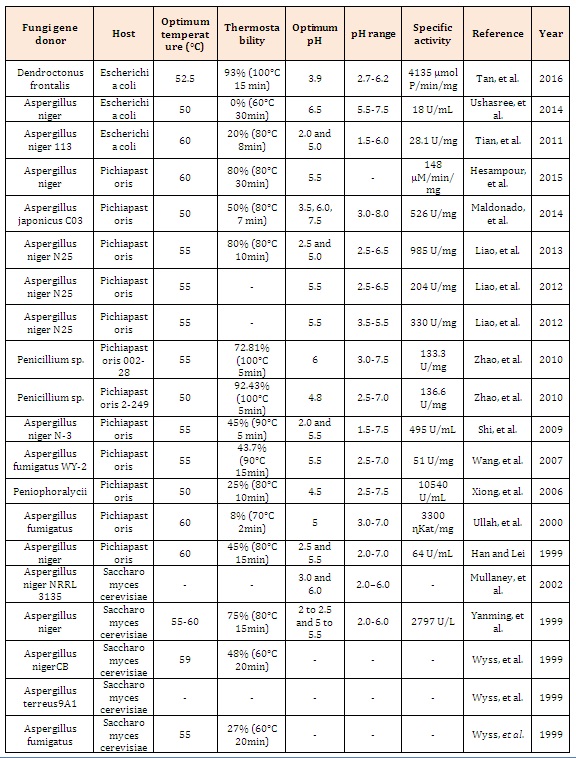
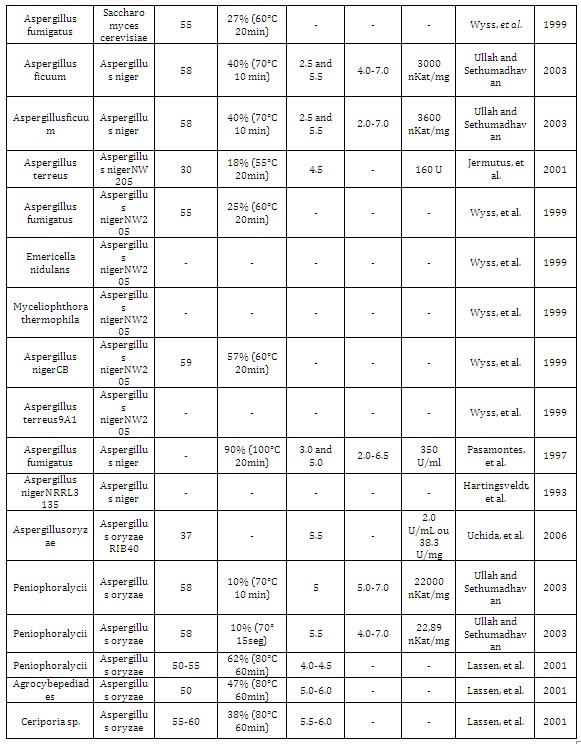
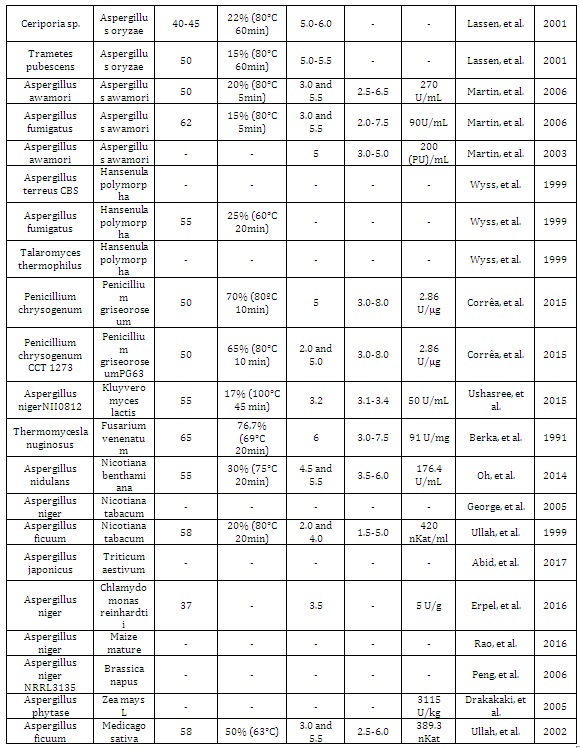
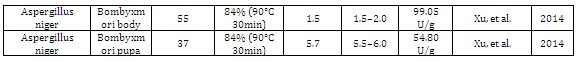
Table 1: Fungal phytases expressed in different hosts and their biochemical properties.