
Citation: Barreto CC, et al. The Cultivable Microbiota Associated with Four Antarctic Macroalgae. J Microbiol Biotechnol, 2017, 2(2): 000122.
*Corresponding author: Cristine Chaves Barreto, Universidade Católica de Brasília, Programa de Ciências Genômicas e Biotecnologia, Brasília – Brazil, Tel: 055 61 3448 9217; Email: criscbarreto@gmail.com
Marine macroalgae are suitable surfaces for bacterial colonization; in fact, this microbiota is important to the cycling of organic material from algal origin. We describe the cultivable microbiota associated with four endemic Antarctic macroalgae: Palmariadecipiens, Himantothallus grandifolius, Desmarestia antarctica, and Ascoseira mirabilis. Among the bacterial colonies obtained on marine agar 71.4% were affiliated to Proteobacteria, 20% to Firmicutes, and 8.6% to Bacteroidetes. Pseudoalteromonas was the only genus present in all samples; otherwise, each macroalga presented a unique microbiota, which was distinct from macroalgae from northern oceans. These results indicated a species-specific interaction between Antarctic macroalgae and its surface microbiota.
Keywords: Antarctic Seaweed; Antarctic Macroalgae; Seaweed Microbiota; Macroalgae Microbiota
The microbial community associated to macroalgae (seaweed) is diverse and complex, differing among algal phyla and species, season, and host age [1]. Interactions between marine macroalgae and microorganisms are very diverse as well; macroalgae provides home and nutrients, a resultant from algal photosynthesis, to associated microorganisms [2]. On the other hand, it was demonstrated that bacteria influence algal growth by producing vitamins [3] or even increasing bio-availability of Fe [4]. Another example is the highly specific association between Vibrio angularum and the green macroalga Ulva in which algal zoospores will only establish on a surface in the presence of certain chemical signals from their bacterial partner [5]. Some authors also reported the essential role of specific bacterial communities for the normal development of the algal host [6,7]. Such intimate association suggests that macroalgae and bacteria interact as a unified functional entity or holobiont [8].
There are few studies on the ecological aspects of the interactions between macroalgae and its microbiota. The majority of the studies describe the bacteria associated with edible macroalgae, especially Pyropia, Laminaria and Undaria that are consumed in Japan, Korea and China as well as Palmariapalmata which is consumed in Ireland. Such studies were directed to identify one algal or human pathogen on themacroalgae [9,10]. The prevalence of the family Flavobacteriaceae was reported previously in studies on the macroalgae from the Gulf of Peter the Great (Sea of Japan) [11-13].
The importance of the macroalgae and their associated microbiota to the carbon cycle in Antarctic oceans may be underestimated.Scientific studies revealed the presence of many species of microorganisms in Polar environments unveiling the important role of Antarctic bacteria in consuming, producing, and sequestering different kinds of compounds. Although macroalgae are abundant in the Antarctic Peninsula, little is known about the diversity of the microbiota associated with macroalgae from cold environments, especially from species endemic to polar environments.
An undergoing research developed in the surroundings of the Brazilian Antarctic Scientific Station – Comandante Ferraz (EACF) showed that the Rhodophyte Palmariadecipiens is the most common macroalga in the Antarctic Peninsula [14]. The estimated biomass produced by deposition of macroalgae casted ashore on the beach of EACF is about 60 kg by lineal meter of shore, with P. decipiens comprising up to 95% of this biomass.The objective of the present work was to survey the bacterial species that inhabit the surface of macroalgae by isolating and characterizing heterotrophic bacteria associated with four common species of marine Antarctic macroalgae.
Material and MethodsLiving individuals of four species of macroalgae were collected in Admiralty Bay, King George Island, in February of 2007. Samples of Ascoseira mirabilis and Himantothallus grandifolius were collected in Demay Point; Desmarestia antarctica in Agat Point; and Palmariadecipiens from areas close to the Brazilian station [14].
Each alga was washed three times in sterile distilled water to remove bacteria that were loosely associated; 10 g of each alga were grinded in sterile seawater. A 100-fold dilution of this mixture was inoculated on Marine Agar 2216 (DIFCO) and the plates were incubated at 10ºC for 7 to 11 days. Bacterial colonies were selected based on morphological characteristics. Cell morphology was checked for purity and uniformity using bright field microscopy after a Gram staining.
DNA of each colony was extracted from 5 mL of liquid culture [15]. The gene for the 16S rRNA was amplified by PCR reaction using the Universal primers 27F and 1401R [16,17]. The nearly-full sequences of the 16S rRNA gene were obainted using the same primers described above. The nucleotide sequences were analyzed using the software Bio Edit [18] and Bacterial affiliation was determined using the Ribosomal Database Project II [19]. Corresponding sequences were deposited in the GenBank under the numbers JQ618815-JQ618847.
Multivariate analyses were applied to summarize the data. A matrix containing the microbiota associated with macroalga species was generated and used for the Detrended Correlation Analysis (DCA), revealing the linear distribution of data (gradient size < 4.0), which was further analyzed by Principal Component Analysis (PCA) [20]. The multivariate analyses were carried out using Canoco 4.5 [21].
Results and DiscussionThirty three cultivable bacteria associated with Antarctic macroalgae were isolated. Seven isolates (21.2%) were Gram-positive cocci and 26 (78.8%) Gramnegative bacilli. The analysis of the 16S rRNA gene revealed that they belonged to three Bacterial phyla: Firmicutes, Bacteroidetes and Proteobacteria (Table 1)
Proteobacteria was the most abundant and diverse Bacterial phylum (71.4%) found among the isolates; they were either affiliated with the Gamma Proteobacteria (Pseudoalteromonas, Halomonas, Cobetia, Marinomonas, Colwellia, Pseudomonas, and Psychrobacter) or to the Alpha Proteobacteria (Sulfitobacter). Phylum Firmicutes was the second most represented (20%); the sequences were affiliated with the genera Aerococcus or Planococcus, representing the Gram positive bacteria initially observed. Only three bacteria were affiliated with the Phylum Bacteroidetes (8.6%), these bacteria were related to the genera Maribacter, Polaribacter, and Winograskyella (Table 1). Previous studies revealed that these phyla are common in polar marine environments such as: sea ice [22], seawater [23], and macroalga from the Gulf of Peter the Great [11,13].
Although some of the genera were already observed in association with marine macroalgae from northern oceans, the composition of their microbiota is distinct from the Antarctic macroalgae [11,24]. In addition, the genera Aerococcus, Polaribacter, Winogradskyella, and Colwellia were for the first time observed on such surfaces. It is possible that the differences observed on cultivable microbiota from Antarctic and non-Antarctic macroalgae may be due to differences in culture media or cultivation techniques, since the choice of culture medium favors the growth of specific genera.
The genus Pseudoalteromonas was observed in all four Antarctic macroalgae; over 61.5% of the isolates associated with A. mirabilis belonged to this bacterial genus. Except for the genus Pseudoalteromonas, each macroalgae species studied presented a distinct bacterial composition, which was further validated statistically by PCA; that showed differences in the microbiota of the four macroalgae species analyzed (Figure 1). This result suggests a species-specific association between macroalga and microbiota. This observation is in agreement with previous studies comparing the cultivable microbiota, which indicated a species-specific microbial community on non-Antarctic algal species from the Sea of Japan [11] and Tuandao Bay, China [24].
Although molecular analysis applied on the metagenome may provide a better representation of the microbial richness from an environmental sample, it is not free from technical biases [25]. One advantage of classic culture techniques is the possibility of further studies on interactions between organisms and the screening of novel bacterial products for biotechnological applications [1]. Therefore, the present study provided not only information about the cultivable heterotrophic bacteria on Antarctic algal surfaces, but also allows for further investigation on the nature of this interaction.
AcknowledgementsThis research was funded by the Brazilian National Counsel of Technological and Scientific Development – CNPq. Rubens T. D. Duarte received scholarship from Fundação de AmparoàPesquisa do Estado de São Paulo (FAPESP - grant #2012/11037-0). This study was made possible with the logistic support from the Brazilian Antarctic Programme (PROANTAR). It was included in the API activity 403 “MIDIAPI Microbial Diversity of Terrestrial and Maritime ecosystems in Antarctic Peninsula” under the 2007-2008 International Polar Year activities of Projects MERGE (Microbiological and Ecological Responses to Global Environmental Changes in Polar Regions), CAML (Census of Antarctic Marine Life), and SCAR MarBIN (SCAR-Marine Biodiversity Information Network). We thank Dr. Fernando Dini Andreote and Dr. Ederson da Conceição Jesus for the statistical support.
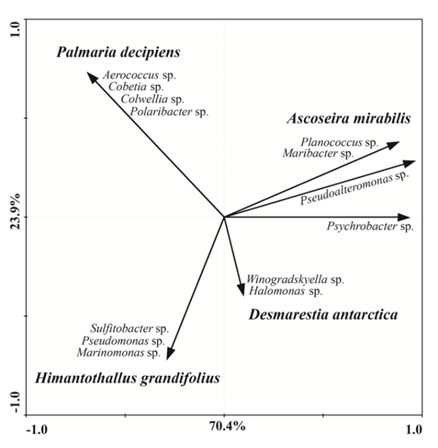
Figure 1: PCA showing bacteria associated with each species of macroalga. The first two ordination axes explained 94.3 % of the total variance. Macroalgae species are presented in bold italic and their correspondent microbiota in italic.
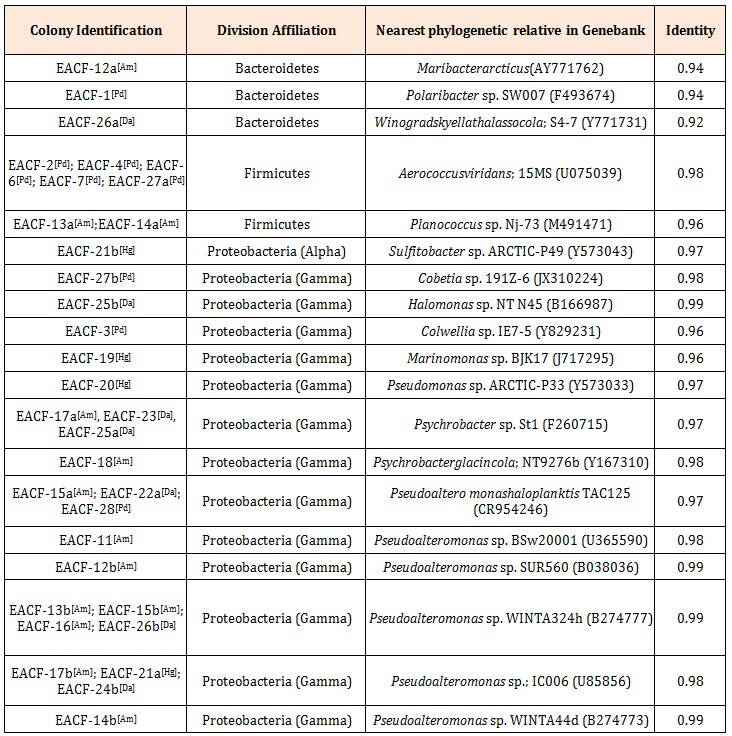
Table 1: Affiliation of the isolated bacteria associated with macroalga according to the 16S rRNA gene phylogeny. Letters inside brackets indicate the macroalga from which this bacterium was obtained: Am: Ascoseira mirabilis, Hg: Himantothallusgr andifolius, Da: Desmarestia antarctica, and Pd: Palmariadecipiens