
Citation: Ates I, et al. Possible Effects of Paraoxonase 1 Q192r Polymorphism on Serum Paraoxonase Activities of a Group of Type 2 Diabetes Mellitus Patients. J Pharm Res 2017, 1(4): 000122.
*Corresponding author: Ilker Ates, Ankara University, Faculty of Pharmacy, Department of Toxicology, Tandogan 06100, Ankara, Turkey, Email: ilkerates976@gmail.com
Paraoxonase (PON) 1 is an antioxidant enzyme a member of PON family held accountable for the protection against several diseases, exposures and xenobiotics like paraoxon. It has been shown that genetic and phenotypic changes in PON1 concentration evoke some diseases such as Diabetes Mellitus (DM). The recent studies indicated in both Type 1 and Type 2 DM patients; serum PON1 activity and concentrations are diminished. In the light of this information, we carried out this study to evaluate lipid profiles and possible relationship between PON1 activity and PON1 Q192R polymorphism in a group of Turkish Type 2 diabetic patients. Due to our experimental data, the PON activity of the patients with minor RR variant is significantly higher than patients with QQ and QR variants (p<0.01). To conclude, we observed PON1 192RR genotypes are relevant to enhanced PON activity than the other variants. This situation may be interpreted as more protective to lipid peroxidation.
Keywords: Pon1; Serum Pon1 Activity; Pon1 Q192r Polymorphism; Lipid Peroxidation; Type 2 Dm
The paraoxonase (PON) gene family has three member enzymes (PON1, PON2 and PON3) displaying antioxidative features especially in blood circulation. Among these members, PON1 remains the most remarkable and studied enzyme [1].
PON1 is a Ca+2-dependent serum esterase first identified for its capacity of hydrolyzing pesticides and organophosphates, containing paraoxon. PON1 is a 43–45 kDa glycoprotein, expressed in several tissues [2], but it is actually synthesized by the liver and following the synthesis it circulates within high-density lipoprotein (HDL) particles [3]. It became the focus of more dense researches, due to its evident capacity of protection of low-density lipoproteins (LDL) versus oxidative stress, decreasing the generation of macrophage foam cells and hindering the development of atherosclerosis [4]. PON1 gene polymorphisms have been linked with several diseases, comprising Parkinson’s disease, psoriasis, inflammatory bowel disease, coronary heart disease and type 2 diabetes [5-11].
PON1 gene is located at q21-q22 on the long arm of chromosomes in humans [12]. A common amino acid at location 192 (glutamine [Q allele]/arginine [R allele] has been defined in the coding sequence of PON1 of human [13]. A possible polymorphism at this location has a substrate-dependence character. Some substrates like fenitroxon and paraoxon are hydrolyzed rapidly by R allele while others including phenyl acetate is hydrolyzed at the same ratio by both alleles. Conversely, sarin, soman and diazoxon substrates are hydrolyzed faster by the Q allele [14].
Similarly the other diseases, inflammation via enhanced oxidative stress plays an important role in DM aetiology. Diminished PON 1 activity is related with the augmented lipid peroxidation and this may be the major determinant in the development of diabetes complications. Various recent studies showed that there is a significant decrease in PON1 activity of diabetic patients in contrast with controls [15-18]. The underlying mechanism of this fact is not absolutely understood yet; several determinants are believed to take role in this alteration. The first determinant is that the alteration in antioxidant capacity of PON1 through activity drop speeds up the oxidative stress and therefore lipid peroxidation may conduce to the breakdown of vascular wall [19]. The second one is that the grossly glycation of the enzyme contributes to the reduction of the PON1 activity in disease [20]. This research is designed to evaluate the lipid profiles and the possible relationship between PON1 activity and PON1 Q192R polymorphism in a group of Turkish Type 2 diabetic patients.
Materials and MethodsA control group of 50 healthful individuals (21 male and 29 female) without any signs of Diabetes mellitus and 100 patients (56 male and 44 female) diagnosed of Type 2 diabetes were counted as study groups. Individuals hospitalized in Diabetes Clinic of Ankara Training and Research Hospital diagnosed of Type 2 diabetes with regard to the criteria regulated by the World Health Organization (WHO, 1999) was picked [21]. All study subjects gave a written informed consent. Also this research was approved by the Ethics Committee of Ankara University, Faculty of Medicine and Ankara Training and Research Hospital.
PON Activity AnalysisSerum PON1 activity was detected due to the methods defined by Eckerson, et al. and Mackness, et al. [22,23]. Activity was calculated by appending serum preincubated with 1x10-5 mol serine to Tris/HCl buffer (0.1 mol, pH 8.0) including 2 mmol CaCl2 and 2 mol paraoxon (Sigma). The formation ratio of p-nitro phenol was identified at 25ᵒC with a spectrophotometer at 405 nm. The method defined by Eckerson, et al. was employed for the calculation of arylesterase activities. Activity was calculated by appending serum preincubated with Tris/HCl buffer (9 mM, pH 8.0) including 1 mM phenyl acetate and 0.9 mM calcium chloride. Activity was calculated with a spectrophotometer at 270 nm at 25ᵒC.
Preparation of DNA and GenotypingPromega Wizard Genomic DNA Purification Kit was used according to the manufacturer’s directive for DNA isolation. DNA specimens were stocked at -20ᵒC until the genotyping experiments. PON1 genotype was identified by PCR-RFLP (Polymerase chain reaction-restriction fragment length polymorphism) technique in our laboratory. Primers used for the 192 polymorphism was: 5’-GTATGTTTTAATTGCAGTTTGAA-3’ (forward) and 5’- TGAAATGTTGAT TCCATTAGCAA-3’ (reverse). Hsp92II restriction enzyme was used in RFLP technique.
Other Laboratory CalculationsCholesterol, fasting glucose and triglyceride concentrations were calculated by using traditional methods (Abbott Spectrum Auto Analyzer following precipitation with dextran sulphate/magnesium). Friedewald’s formula was performed for calculating LDL cholesterol. HbA1c was calculated by a latex agglutination inhibition rate assay with DCA 200 analyzer.
Statistical AnalysesThe 16th version of SPSS software (SPSS Inc., USA) was performed. Clinical laboratory data were expressed as means ± SD. Student’s t-test was used for mean values and χ2 test (Fisher Exact Test when appropriate) was used for evaluation of the possible significant associations between polymorphic variants and disease situation. Odds ratios (OR) with 95% confidence intervals (CI) were also measured. With the help of linear regression, the associations between the genotypes and PON activity were identified. P< 0.05 was noted as statistically significant.
ResultsThe demographic data of the all study subjects were demonstrated in Table 1.
It was clearly seen that triglycerides, VLDL-cholesterol, fasting glucose, systolic blood pressure, diastolic blood pressure and HbA1c levels of the patients were significantly above from those of controls. On the other hand, serum arylesterase and PON activities of the patients were lower compared to controls but these are not statistically significant.
In Table 2, Q192R genotype and allelic frequencies of the patients were given. A significant difference was not seen between the gene frequency for PON1 Q192 R polymorphism in both controls and patients due to the χ2 test.
Table 3 demonstrates the association between Q192R variant and PON activity, lipid profiles and HbA1c. According to our data this polymorphism had an important effect on the activity of PON. In both study groups, RR genotype showed the highest PON 1 activity while the QQ genotype showed the lowest. On the contrary, we found no association between the polymorphism and the plasma lipid, cholesterol and HbA1c grades.
DiscussionWe aimed to evaluate the lipid profiles and the possible relationship between PON1 activity and PON1 Q192R polymorphism in a group of Turkish Type 2 diabetic patients in this research. Due to our findings, there was no any significant allelic distribution of PON 192 genotypes between the study groups. These results show similarity to the findings of Agachan et al., but on the contrary to Flekac, et al. study results [24,16]. We observed the decrease of PON 1 activity in patients compared to controls like other researchers Karabina, et al. Flekac, et al. however this decrease is not significant compared to the researchers discussed overhead [25,16].
It is known that PON 1 gene polymorphisms may affect the versatility of enzyme activity. Low activity diminishes the capacity to hinder the generation of lipid peroxide with consequent momentum of oxidative stress leading to the possible enhanced lipid peroxidation in diabetes. Due to our data, PON1 Q192R polymorphism was related with the enzyme activity, that augmented in the row of QQ < QR < RR genotypes. In both study groups, QQ genotypes have the lowest enzyme activity whereas the RR has the highest. Our findings encourage the opinion that lipid peroxidation versus oxidation by PON 1 might be lowered in patients because of lower activity of the enzyme declared by researchers including Agachan, et al. and Flekac, et al. [24,16].
The reason for diversity among the studies searching PON 1 genotype distribution might be related with variances between ethnic populations [26]. The research incoherency in the relationship between PON genotypes and diabetes might be on account of the boundaries of traditional retrospective and case-control researches forwhy sampling bias has to be kept in mind.
ConclusionThe data of our research recommend that PON enzyme activity is affected by genetic inequality in Turkish patients with diabetes and control individuals. We observed PON 1 192RR genotype is tightly related with the enzyme activity compared with the others. This situation may generate a protection to lipid peroxidation.
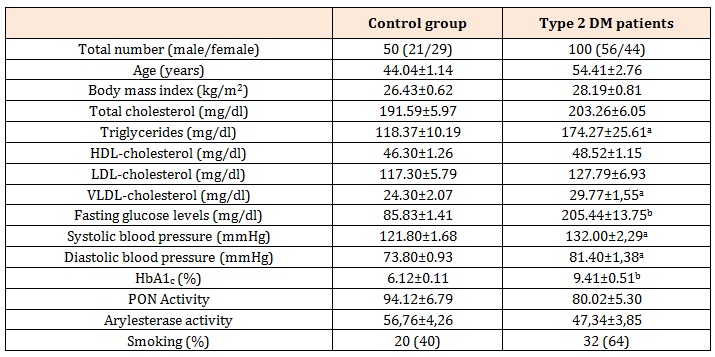
Table 1: Demographic details of the study groups.
HDL: high-density lipoprotein, LDL: low-density lipoprotein.
a p<0.05 compared with controls
b p<0.01 compared with controls
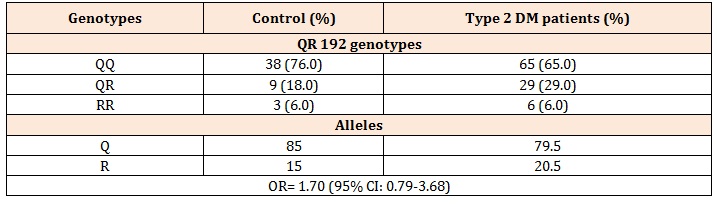
Table 2: PON1 Q192R genotype distribution and allele frequencies in the study groups.
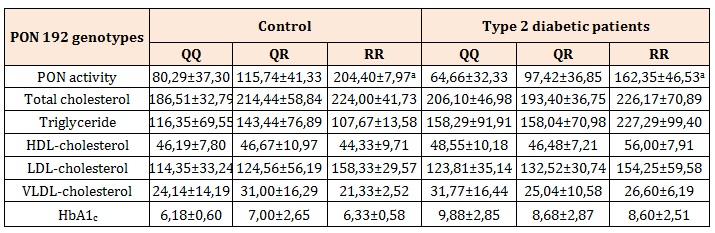
Table 3: The relationship between Q192R variant and PON activity, lipid profiles and HbA1c.
a P<0.01 compared to the PON1 activities of QR and QQ variants in both study groups