
Citation: Chiou RYY, et al. PCR Detection of Botryosphaeria rhodina as a Biotic Elicitor in Enhancement of Trans-Resveratrol and Trans-Piceid Biosynthesis of Peanut Kernels during Germination J Agri Res 2016, 1(2): 000111.
*Corresponding author: Robin YY Chiou, Department of Food Science, National Chiayi University, Taiwan, Tel: +8865- 2717613; Email:rychiou@mail.ncyu.edu.tw
A fungus involved in enhancement of trans-resveratrol and trans-piceid biosynthesis of peanut kernels during germination was isolated and molecularly identified by PCR as Botryosphaeria rhodina. When peanut kernels were surface-disinfected, 2-d incubated for germination and artificial colonized with the isolated B. rhodina at 20 or 30oC for 96 h, trans-resveratrol and trans-piceid contents of the kernels increased substantially with time of artificial colonization. The interaction of B. rhodina with peanut kernels during germination in elicitation of resveratrol and piceid biosynthesis is obvious. For development of a rapid PCR procedure in screening B. rhodina isolates, which are widespread in nature, a specific primer set was designed based on the internal transcribed spacer (ITS) region of ribosomal DNA and used for PCR detection of the morphology-presumptive isolates of B. rhodina, a targeted 424-bp fragment was exclusively detected for the B. rhodina isolates. The fragment was also detected when the genomic DNA’s extracted from peanut kernels after colonized with B. rhodina were used as template for PCR amplification. The quantity of the amplified fragment increased with an increase of time of colonization.
Keywords: Resveratrol; Piceid; Peanut; Elicitor; Biosynthesis; Botryosphaeria rhodina
Resveratrol (3,5,4’-trihydroxystilbene), a plant secondary metabolite, has been demonstrated to confer potent effectiveness in chemoprevention of chronic diseases including cardiovascular diseases, cancers and even extension of lifespan [1-8]. Extensive research interests have been addressed on biotic and/or abiotic elicitation of plants, such as edible crops with particular interest, to enhance resveratrol biosynthesis [9-13]. Resveratrol has been detected in peanut kernels, roots, leaves, pods, peanut-related foods, sprouts, cultured callus and root mucilage [14-26]. Peanut is genetically capable of biosynthesizing resveratrol and other stilbenoids. However, resveratrol contents of the harvested peanut kernels and peanut products are considerably low [16-18]. Enhanced biosynthesis of resveratrol and other stilbenoid phytoalexins are necessarily elicited by external challenges including physiological stress, wounding and microbial infection [13, 27-31]. In our laboratory, enhancement of resveratrol biosynthesis during germination of peanut kernels in preparation of peanut sprouts has been observed [24]. In addition to resveratrol, biosynthesis of other stilbenoids including arachidin-1, arachidin-3 and isopentadienyl resveratrol were also enhanced by germination, slicing and incubation with artificial aeration [26]. These observations revealed that peanut could be developed as a potent bioreactor in biosynthesis of resveratrol and other bioactive stilbenoids for further related product development.
Germination of seeds is a crucial step in generation of a new plant. During germination, a complex and sophisticated set of biochemical and physiological reactions are undergoing through precisely controlled mechanisms. Pathogen infection may cause severe damages on host plants, while a mild- or non-pathogenic infection may only elicit phytoalexin biosynthesis. In our recent experiments, a fungal contaminant has been frequently observed during peanut kernel germination. As a preliminary investigation, the resveratrol contents of those infected peanut cotyledons were much higher than those of the normally germinated ones. This has raised our attention and interest to isolate the fungus for further investigation. Thus, the isolate was first subjected to molecular identification as Botryosphaeria rhodina RCYU 30101. For elicitation, novel peanut kernels were surfacedisinfected and subjected to germination and artificial colonization by the isolate. Subsequent extraction of the kernels with methanol solution for HPLC quantifications of trans-resveratrol and trans-piceid contents were conducted.
In nature, Botryosphaeria isolates are widespread association with agricultural produces. The asexual state of B. rhodina is also named as Lasiodiplodia theobromae. A rapid molecular differentiation of B. rhodina from other molds is of practical importance. For development of a rapid PCR procedure in screening B. rhodina isolates, a specific primer set was designed based on the internal transcribed spacer (ITS) region of ribosomal DNA and applied for PCR differentiation of B. rhodina. By the developed PCR procedure, the artificially colonized B. rhodina in the peanut kernels was detected. In addition, some morphology-assumptive Lasiodiplodia sp. isolates originated from local fruits of mango, papaya and guava were also subjected to the developed PCR detection.
Materials and Methods PeanutsPeanuts of Tainan 14 (Arachis hypogea L., a Spanish cultivar) were used in this experiment. The harvested, sun dried, hand shelled and graded peanut kernels were sealed in high-density polyethylene (PE) plastic bags and stored at –25°C for use.
Peanut germination and isolation of fungal contaminantsPeanut kernels were imbibed with tap water, drained and incubated for germination and sampled for HPLC quantification of resveratrol content following the procedure described previously [22]. As observed, transresveratrol contents of the germinated kernels colonized with a white mold were much higher than kernels colonized with other molds or the normally germinated kernels. For isolation of the fungal contaminants, the cotyledon splits obtained from mold colonized kernels were subjected to surface disinfection by stepwise immersing into 1% NaOCl solution for 1 min, 75% ethanol for 1 min, rinsed with sterile water for 4 times and aseptically cut into slices. After deposition of the slices onto potato dextrose agar (PDA) plates for incubation at 28°C for 7 d, pure isolates were further stripped and subcultured onto new PDA plates.
From each colony, a small part of mycelium was aseptically removed with a loop and incubated on a PDA plate for pure colony formation. Then, the colonies were subjected to morphological examination and classified according to the identification criteria of imperfect fungi [32]. Pycnidiospore morphology of the mature spores produced by the isolates was examined microscopically. An isolate bearing pycnidia with ovary shape, brown pigmented, septum divided (2 cells) with striate and its pycnicia clustering onto a stroma was identified as a morphology-presumptive Lasiodiplodia sp. (the asexual state ofBotryosphaeria sp.).
DNA extraction and PCR amplificationThe procedures reported by Yoon et al. [33] and Chen et al. [34] were followed and modified to extract the genomic DNA from each fungal colony. Briefly, a small amount of mycelial mat was taken and deposited onto a sterile mortar, quick frozen with liquid nitrogen and ground with a sterile pestle into powder, which was further ground with 400 μL of lysis buffer (API buffer). Then, the solution was transferred into a new 1.5 mL microfuge tube offered by a commercial kit (DNeasy Plant Mini Kit, Qiagen GmbH, Hilden, Germany) and following the manufacturer’s procedure for DNA extraction. DNA purity and concentration of the extracted solutions were determined spectrophotometrically (GeneQuant II, Pharmacia, Cambridge, UK). A universal primer set shown in Table 1 was used to amplify fragments of the 18S rDNA, ITS1, 5.8S rDNA and ITS2 region [30]. For PCR amplification, 25 μL of PCR reagents including 1 μM of each primer, 0.2 mM for each of four deoxyribonucleotides and 1 unit of AmpliTaq DNA polymerase in 1 x PCR buffer were conducted. The reactions were performed by using GeneAmp PCR System 2400 (Applied Biosystem, Norwalk, CT, USA).
Sequencing of the PCR productsThe PCR products were directly ligated with pGEM®-T Easy vector (Promega Co., Madison, WI, USA) and the ligation mixtures were then transformed into competent cell of Escherichia coli JM109 according to the manufacturer’s instructions. DNA sequence was determined by an auto sequencer of ABI 3730 (Applied Biosystem). The sequences were compared to the GenBank database using BLAST program of NCBI.
Rapid PCR detection and screening of B. rhodina isolatesFor development of a rapid PCR procedure in screening B. rhodina isolates, a specific primer set was further designed based on the internal transcribed spacer (ITS) region of ribosomal DNA to amplify a 424-bp fragment (Table 1). For PCR amplification, 20 μL of PCR reagents including 2 μL of DNA template, 1 μL (10 M) of each primer, 4 μL of 5 x Taq Master Mix (Protech Tech. Enterprize Co., Ltd., Taipei, Taiwan) and 4 μL deionized water were subjected to PCR multiplication by GeneAmp PCR System 9700 (Applied Biosystem). The PCR reactions are listed in Table 1. The PCR products were separated by gel electrophoresis in 1.5% at 100 V and a distinguished DNA fragment was visualized under UV and photographed (Polaroid GelCam and Polaroid 667) after ethidium bromide staining. The sequence of the fragment was determined as that described above.
For rapid detection of B. rhodina by PCR, 4 morphology-presumptive isolates of Lasiodiplodia sp. (a sexual state of Botryosphaeria sp.) and 7 other fungal isolates collected from peanut kernels during germination, and 10 other mold cultures of Aspergillus oryzae BCRC 30120, Asp. oryzae BCRC 30123, Asp. oryzae BCRC 33705, Asp. flavus BCRC 30119, Asp. flavus BCRC 30166, Asp. parasiticus BCRC 30160, Rhizopus microsporus var. oligosporus BCRC 31996, Rhizomucor miehei BCRC 33081, Penicillium digitatum BCRC 31556 and Asp. niger RCYU 30111 were subjected to genomic DNA extraction and PCR detection with the primer set that described above. For broadening assessment spectrum in identifying fungal contaminants, 20 Lasiodiplodia sp. isolates collected from local mango, papaya and guava were subjected to genomic DNA extraction and PCR detection as that described above.
Mycelial inoculum and artificial colonization of the germinating peanut kernelsA loopful of mycelium of B. rhodina RCYU 30101 was inoculated into yeast malt broth (YMB) (50 mL in each 125 mL flask) and incubated at 30°C (100 rpm) for 48 h. After incubation, the flasks containing mycelial suspension were stored at 4°C and used as an inoculum.
For artificial colonization, novel peanut kernels were surface disinfected under aseptic condition by stepwise immersing the kernels into each of 1% NaOCl and 75% ethanol for 1 min. Then, the kernels were rinsed with sterile water 4 times and immersed in sterile water for 3 h. After draining the water, 35 kernels were deposited into a sterile flask (500 mL), capped with aluminum foil and incubated at 20 and 30 oC for germination (Hipoint 747FH, Hipoint Co., Kaohsiung, Taiwan).
After 2 d of incubation, 1 mL of the B. rhodina mycelial suspension was dropped and distributed evenly onto each surface of the germinating kernels. The flasks, after artificial inoculation, were further incubation at 20 and 30oC for germination. During additional incubation for 96 h, 4 kernels were randomly sampled from the flask at 0, 17, 24, 48, 72 and 96 h for resveratrol determination and genomic DNA extraction.
Extraction of trans-resveratrol and trans-piceid for quantification and genomic DNA for PCR detection of B. rhodinaCotyledons from each of the germinating kernels (on single kernel basis) were freeze-dried (Lyvotac GT2, FinnAqua, Heraus, Germany), pulverized and stored at -25°C for later use. Twenty mg of the freeze-dried powder was deposited into a microfuge tube (1.5 mL) provided by a commercial kit (DNeasy Plant Mini Kit) and mixed with 1.0 mL 80% methanol. After grinding with a sterile cellular pestle, the tubes were centrifuged and the supernatants were subjected to HPLC quantification of trans-resveratrol and trans-piceid (glucoside of resveratrol) contents following the procedure reported by Chang et al. [26]. Authentic trans-resveratrol and transpiceid (Sigma Co., St. Louis, MO, USA) were run concurrently as references for identification and quantification.
For subsequent extraction of genomic DNA from the B. rhodina colonized peanut kernels after methanol extraction and removal of supernatant for HPLC analysis of resveratrol, the pellet in a microfuge tube (DNeasy Plant Mini Kit) was replenished with 400 μL AP1 buffer and quick frozen with liquid nitrogen [34]. After grinding with a sterile cellular pestle, the manufacturer’s procedure for a commercial kit (DNeasy Plant Mini Kit) was followed for DNA extraction. Concentration and purity of the extracted DNA were determined as that described above. Then, the DNA extracts were subjected to rapid PCR detection and electrophoresis following the procedures described above.
StatisticsMean of determinations with standard error for transresveratrol and trans-piceid contents and analysis of variance (ANOVA) conducted by SAS are reported.
Results Molecular identification of B. rhodina isolated from the germinating peanut kernelsGenomic DNA’s were extracted from the isolated fungal contaminants observed as biotic elicitors of resveratrol biosynthesis during peanut germination. After PCR amplification with universal primers of ITS1 and ITS4, sequences of an amplified 537-bp DNA fragment and a referenced B. rhodina with accession number AY 754002.1 in GenBank database are shown in Figure 1. The alignment result showed 100% sequence identity with the reported sequence of B. rhodina which possessed a 751-bp sequence containing partial 18S rRNA, internal transcribed spacer (ITS1), 5.8S rRNA and ITS 2 [36]. Accordingly, the isolate was molecularly identified as B. rhodina, which is classified as Eukaryota; Fungi; Ascomycota; Pezizomycotina; Dothideomycetes et; Chaetothyriomycetes incertae sedis; Botryosphaeriaceae; Botryosphaeria. The asexual state of B. rhodina (Berk. & M.A. Curtis) Arx is also named as Lasiodiplodia theobromae (Pat.) Griff. & Maubl. (= B. theobromae Pat.). This isolate was assigned as B. rhodina RCYU 30101 in our institution.
Elicitation of trans-resveratrol and trans-piceid biosynthesisWhen novel peanut kernels were surface-disinfected, artificially inoculated with the mycelial suspension of B. rhodina RCYU 30101 and germinated at 20 and 30°C for 96 h, changes of trans-resveratrol and trans-piceid contents are shown in Tables 2 and 3. Resveratrol contents increased substantially (P < 0.05) after 24 h of colonization with B. rhodina either at 20 or 30°C (Table 2).
The highest resveratrol levels for the B. rhodina colonized kernels were 316.5 and 354.9 µg/g freeze-dried kernel weight after 96 h of incubation at 20 and 30°C, respectively. For the un-inoculated kernels, resveratrol contents increased from 7.7 to 31.0 µg/g as affected by time and temperature of incubation. As affected by temperature, resveratrol contents of the B. rhodina colonized kernels incubated at 30oC were higher than those of the kernels incubated at 20°C (P < 0.05).
Similarly, a significant effect on enhancement of transpiceid biosynthesis by colonization infection with B. rhodina was observed (Table 3).
For the un-inoculated kernels, trans-piceid contents changed in a limited range from18.7 to 31.5 μg/g. In correlating trans-resveratrol and trans-piceid contents biosynthesized by the kernels (Tables 2 and 3), a simultaneous increase of both trans-resveratrol and trans-piceid contents was not observed.
Rapid detection of B. rhodinaFor rapid detection of B. rhodina by PCR, a specific primer set, BR1 and BR2, was designed based on the sequence of the obtained fragment (Table 1). As applied for PCR reaction, the expected 424-bp fragment was only detected for B. rhodina RCYU 30101 and 3 other Lasiodiplodia sp. isolates (morphology-presumptive B. rhodina) (Figure 2A). No amplified fragment was detected from other tested mold cultures. When 20 Lasiodiplodia sp. isolates collected from local mango, papaya and guava were subjected to genomic DNA extraction and PCR detection, the specific fragment was also detected in all test Lasiodiplodia sp. isolates (Figure 2B).
Screening and detection of B. rhodina from the colonized peanut kernelsAfter the B. rhodina RCYU 30101 colonized peanut kernels were subjected methanol extraction and followed by genomic DNA extraction and PCR, the targeted 424-bp DNA fragment was detected only in the kernels after artificial colonization (Figure 3). In sequencing the obtained fragment, the sequence was 100% identical to the specified target sequence (Figure 1). In addition, intensity of the fragment increased with an increase of time of colonization (Figure 3).
DiscussionBased on the sequence of the amplified fragment of PCR with universal primers of ITS1 and ITS4 (Figure 1), the fungal resveratrol elicitor isolated from the germinating peanut kernels possessed identical sequence (100% sequence identity) to that of B. rhodina (AY 754002.1) listed in GenBank database [36]. Thus, the isolated culture was identified and assigned as B. rhodina RCYU 30101. B. rhodina has a worldwide distribution in the tropical and subtropical regions and occurs on a wide range of plants [37,38]. Botryosphaeria species have been identified and demonstrated as pathogen in association with grapevine decline in western Australia [39]. B. rhodina or named Lasiodiplodia theoromae causes shoot blight of pistachio in California [40] and collar rot of peanut [41].
When a primer set was further designed (Table 1) to amplify a conservative and strain-specific DNA fragment (Figure 1), a specific 424-bp fragment was only detected in B. rhodina RCYU 30101 and 3 other morphologypresumptive B. rhodina (Lasiodiplodia spp.) isolates (Figure 2A lanes 1-4). The fragment was neither detected for other mold cultures isolated from the germinating peanut kernels nor the 10 collected mold cultures, namely, Aspergillus oryzae, Asp. flavus, Asp. parasiticus, Rhizopus microsporus var. oligosporus, Rhizomucor miehei, Penicillium digitatum and Asp. niger (Figure 2A lanes 12- 21).
With an attempt to use the developed PCR to rapid detection of the B. rhodina isolates collected from local mango, papaya and guava, the targeted 424-bp fragment was detected in all 20 morphology-presumptive B. rhodina (Lasiodiplodia spp.) isolates (Figure 2B). As applied the PCR technique for detecting the B. rhodina colonized peanut kernels, the targeted fragment was detected only in the kernels artificially colonized with B. rhodina (Figure 3). The sequence of the fragment was identical to the targeted sequence (Figure 1). As affected by time of colonization, the band intensity increased with time of colonization. Thus, it is undoubted that the kernels have been colonized with the artificially inoculated B. rhodina. Based on the fact that trans-resveratrol and trans-piceid contents of the germinating kernels increased substantially with time colonization, B. rhodina has played a role as a potent biotic elicitor in enhancement of resveratrol biosynthesis.
The highest trans-resveratrol levels of the germinating kernels artificially colonized with B. rhodina RCYU 30101 at 20 or 30°C for 96 h were 316.5 and 354.9 µg/g, respectively (Tables 2 and 3). As compared to the considerably low trans-resveratrol contents for the surface-disinfected kernels after 2-d germination or the un-inoculated kernels, it is obvious that resveratrol biosynthesis has been largely driven by B. rhodina colonization along with kernel germination. This was in agreement with the report of Ingham [14] who observed that cis- and trans-resveratrol formation of peanut hypocotyls was enhanced by fungal infection. As reported by Sobolev et al. [42,43], biosynthesis of resveratrol and its derivatives were stimulated by artificial inoculation of the peanut slices with Aspergillus flavus. Based on the fact that B. rhodina (Lasiodiplodia theoromae) is a plant pathogen known to cause collar rot of peanut [41], infection of the germinating kernels with B. rhodina has provoked a vigorous plant-mold interaction and eventually elicited phytoalexin biosynthesis. In addition to trans-resveratrol, a significant enhancement of transpiceid biosynthesis of the germinating kernels by artificial colonization with B. rhodina was also observed (Table 3). Piceid is the glycoside of resveratrol and confers more hydrophilic characteristics and higher water solubility than does resveratrol. As visualized and manually examined, most of the B. rhodina colonized kernels germinated much slower than did un-inoculated kernels. After 96 h of germination and colonization, they were still viable with rigid texture. The potency to use the kernels as resveratrol source for extraction and purification or for development of bioactive ingredient is noteworthy.
In conclusion, B. rhodina isolated from the germinating peanut kernels has been demonstrated as a potent elicitor to enhance biosynthesis of trans-resveratrol and transpiceid. The resveratrol-enhanced peanut kernels destined for development of some high value-added products deserves further investigations. Besides, investigations on elicitation mechanisms through peanut-mold interaction and product development with the bioactive ingredients are of worth. Based on the fact that B. rhodina is widespread interacted with plants, the developed rapid PCR detection enabling screening and differentiation of B. rhodina from various origins is of merit.
AcknowledgmentWe are grateful to National Science Council (NSC 95- 2313-B415-003), Republic of China for financial support and to Miss Show-Phon Learn for helpful assistance in the laboratory.

Figure 1: Nucleotide sequence alignment of the PCR products; Peanut 1: amplified with universal fungalspecific primers ITS 1 and ITS4; Peanut 2: amplified with Botryosphaeria rhodina-specific primers BR1 and BR2; AY754002: B. rhodina with accession number AY754002 listed in Genebank database.
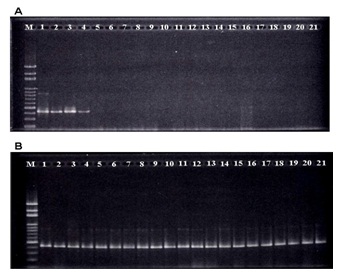
Figure 2: PCR detection of morpholoy-presumptive Botryosphaeria rhodina (Lasiodiplodia theobromae) isolates collected from germinating peanut kernels and 7 other mold cultures; A: M, 100-bp marker; 1, Botryosphaeria rhodina RCYU 30101; 2-4, Lasiodiplodia sp. isolates collected from germinating peanut kernels; 5- 11, other fungal isolates collected from germinating peanut kernels; 12, Aspergillus oryzae BCRC 30120; 13, Asp. oryzae BCRC 30123; 14, Asp. oryzae BCRC 33705; 15, Asp. flavus BCRC 30119; 16, Asp. flavus BCRC 30166; 17, Asp. parasiticus BCRC 30160; 18, Rhizopus microsporus var. oligosporus BCRC 31996; 19, Rhizomucor miehei BCRC 33081; 20, Penicillium digitatum BCRC 31556 and 21, Asp. niger RCYU 30111; B: isolates collected from various local fruits; M, 100-bp marker; lane 1, Botryosphaeria rhodina RCYU 30101; lanes 2, 3, 12 and 17-21, isolates respectively collected from mango; lanes 4-11 and 13-15, isolates collected from papaya and lane 16, isolate collected from guava.
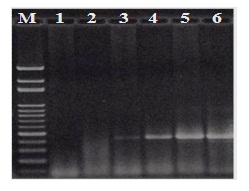
Figure 3: PCR detection of Botryosphaeria rhodina-specific DNA fragments with the genomic DNA’s extracted from surface-disinfected peanut kernels subjected to germination with artificial colonization with Botryosphaeria rhodina RCYU 30101. M, 100-bp marker; 1, peanut kernels before germination (control); 2, peanut kernels after 2-d germination before artificial colonization; 3, 1-d further incubation after artificial colonization; 4, 2-d further incubation after artificial colonization; 5, 3-d further incubation after artificial colonization; 6, 4-d further incubation after artificial colonization.
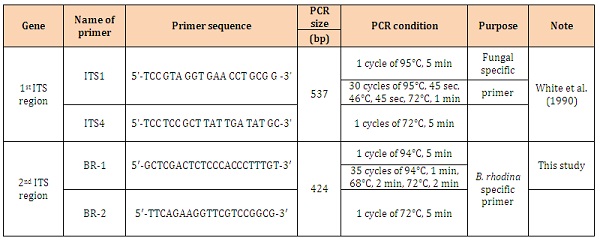
Table 1: Primer sequences and PCR conditions for each primer set.
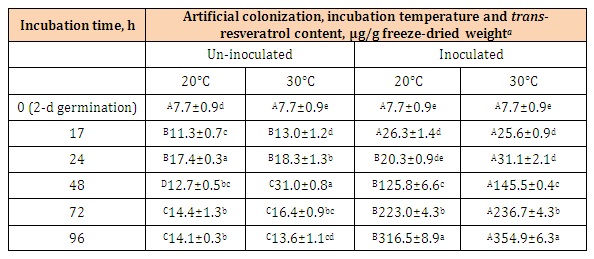
Table 2: Changes of trans-resveratrol contents of the novel peanut kernels (Tainan 14) subjected to surface-disinfection, germination for 2 d and artificial colonization with Botryosphaeria rhodina mycelial suspension and further incubation at 20 and 30°C for 96 h.
aMean of determinations with standard error (n = 3).
A-DData bearing different superscript letters in the same row are significantly different (P < 0.05).
a-eData bearing different superscript letters in the same column are significantly different (P < 0.05).
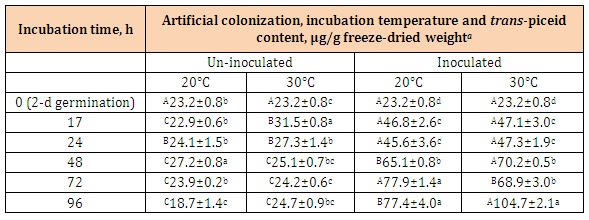
Table 3: Changes of trans-piceid contents of the novel peanut kernels (Tainan 14) subjected to surface-disinfection, germination for 2 d and artificial colonization with Botryosphaeria rhodina mycelial suspension and further incubation at 20 and 30°C for 96 h.
aMean of determinations with standard error (n = 3).
A-CData bearing different superscript letters in the same row is significantly different (P < 0.05).
a-dData bearing different superscript letters in the same column is significantly different (P < 0.05).
Chat with us on WhatsApp