
Citation: Uzochukwu IC, et al. Ending the Ebola Virus Scourge: A Case for Natural Products. J Pharm Res 2016, 1(1): 000105.
*Corresponding author: Uzochukwu Ikemefuna C, Department of Pharmaceutical and Medicinal Chemistry, Faculty of Pharmaceutical Sciences, Nnamdi Azikiwe University, PMB 5025 Awka 420281, Anambra State, Nigeria, Email: ic.uzochukwu@unizik.edu.ng
We investigated the binding affinities of antiviral of ethno medicinal origin to five Ebola viral proteins by in silico molecular docking simulations. One hundred and seventy three compounds were obtained from literature, extracted from ZINC® database as mol2 files and further prepared for docking simulations using Auto Dock tools v. 1.5.6. Five Ebola drug targets (VP24, VP30, VP35, VP40 and NP) were obtained as pdb files from the Protein Data Bank and further prepared for docking simulations using both Chimera v. 1.8.1 and Auto Dock tools v. 1.5.6. Virtual screening and docking simulation experiments were performed using AutoDockVina 4.0 on a Linux platform. Docking results were analyzed using PyMol v.0.99r c6. At least five compounds (robustaflavone, cepharanthine, corilagin, hypericin and theaflavin) were identified as frontrunner antivirals with concurrent binding affinities on the five tested Ebola viral proteins. Binding free energies ranging between -6.225 to -8.675kcal/mol were obtained for the compounds. Compounds are recommended as experimental antivirals of ethno medicinal origin with possible anti-Ebola activities. In vitro and clinical trial investigations into the anti-Ebola activities of these compounds are recommended.
Keywords: Antiviral; Molecular docking; Ebola virus disease; Ethno medicine; Virtual screening
Ebola Virus Disease (EVD) is a hemorrhagic fever caused by Ebola virus belonging to the family filovirideae. In year 2014, West Africa witnessed the longest, largest and most widespread outbreak of EVD. The mortality rate of the recent outbreak of EVD was reported at 30-90 % [1]. The disease caused more than 10,000 deaths from over 25,000 cases [2]. As a result, the WHO declared the epidemic a public health emergency of international concern.
Presently, no medicines or vaccines are proven to be effective for the treatment of EVD. However, a number of experimental drugs and vaccines are under clinical trial in several countries. In the absence of nothing, some physicians have reportedly used some antiviral drugs empirically with some success. Japan promised supplying Nigeria with the antiviral, favipiravir, in the wake of the EVD outbreak in Nigeria. That donation was rejected by the Nigerian Government for undisclosed reasons. Favipiravir was approved by Japan's health ministry for use against influenza. As a result of absence of proven medicines or vaccines, management of the disease was hinged on supportive care with rehydration etc.
Resource poor countries are unlikely to defeat the threat of EVD on their own. As long as EVD persists in these countries, no nation can be said to be free from the risk of EVD. Weak health infrastructure and poverty may remain as major obstacles to the control and elimination of EVD in these countries.
Ebola virus has also been classified as a class A bioterrorism agent. It is among the worst known bioterrorism agent known to man. The implications of bioterrorism with such an agent are many. With Ebola virus in the hand of the ‘bad guy’ without any definitive antidote, its limit of evil could be unequalled.
Experimental investigations of the Ebola virus are hampered by a number of factors. Special facilities and care, the so-called class four bio safety laboratories is a key requirement. This level of bio safety laboratory is non-existent in many developing countries where EVD is ravaging. It is this special difficulty in handling the Ebola virus that lends voice to computational approaches in the discovery of anti-Ebola drugs. We do not need to physically manipulate the virus, while finding answers to the essential scientific questions. The puzzle can indeed be effectively addressed from a distant. This is the gap that computational modeling and simulations can fill.
A number of antivirals and even other pharmacological agents have shown some activities against the Ebola virus in vitro [3]. Some physicians have reported some utility of these agents especially in the absence of proven remedies. We sought to find out if any of these compounds could in fact come to the rescue of EVD. Notably, favipiravir, zanamivir, brincidovir, oseltamivir and clomiphene are claimed to inhibit the Ebola virus. They have therefore been recommended as experimental anti-Ebola drugs.
Most antiviral drugs of synthetic origin are associated with the development of viral resistance, side effects, recurrence and viral latency [4]. The development of drug resistance to these experimental antiviral therefore remains an obvious challenge. It is our postulation that multi-target inhibition answers to the question of resistance. Time shall tell whether this postulation holds true in this case. Part of our current inquiry therefore revolves around the concept of multi-target inhibition of relevant viral targets.
Not minding the difficulty in handling Ebola virus in the laboratory, computational investigations remain feasible and realistic for a number of reasons. First, the Ebola virus is relatively simple, deploying just seven proteins. Any agent that effectively binds to some of the proteins can be trusted to arrest the molecular machinery of the Ebola virus. Secondly, the availability of 3D structures of proteins on the PDB is providing scientists with ready study tool [5]. The ability of scientists to freeze the molecular conformation of Ebola viral protein target is essential in rational design of specific inhibitors. A properly designed inhibitor will hardly miss its target. This is our goal.
The chemical diversity of ethno medicinal compounds can be exploited in the search for novel compounds with anti-Ebola activities. Natural products are known to express unique or novel pharmacophores as part of their survival strategies. Numerous studies have identified antiviral compounds of ethno medicinal origin [4]. It is possible that some of these antiviral of ethno medicinal origin have anti-Ebola activities. This is the crux of our investigations.
Antiviral of ethno medicinal origin usually would have complementary and overlapping mechanism of action, either inhibiting viral replication, or viral genome synthesis. The development of antiviral of natural origin, especially for viral infections for which no approved medication is available, appears to be an urgent necessity. We are also encouraged by the fact that the pathway for introducing a natural product into the clinical armamentarium is less rigorous, moreso for a disease for which no defined treatment course is known. The discovery of an anti-Ebola drug would be of immense benefit to military personnel, health workers, travelers and tourists, who represent the at-most risk group. The study is aimed at identifying antiviral of ethno medicinal origin that could find application as anti-Ebola drugs.
MethodologyBioinformatics mining of the Protein Data Bank (PDB) was done in order to identify Ebola proteins whose 3D structures have been deposited. The 3D atomic coordinates of five identified Ebola virus proteins (VP24, VP30, VP35, VP40 and NP with PDB codes of 4M0Q, 2I8B, 4IBG, 1ES6 and 4QB0 respectively) were obtained from PDB and prepared for molecular docking simulation using UCSF Chimera 1.9 [6] and AutoDockTools 1.5.6 [7, 8]. Briefly, all duplicate chains and hetero molecules were deleted and polar hydrogens added. Grid box sizes, centers and exhaustiveness were assigned to the proteins at 1.0 Å as shown in Table 1. Respective pdbqt files were created for molecular docking simulations studies.
Preparation of ligandsThe electronic structures of one hundred and seventy three (173) compounds, identified from literature (4), were extracted from ZINC ® database (9) and prepared for molecular docking simulations using AutoDockTools 1.5.6 (7, 8). Briefly, rotatable bonds were assigned for the ligands, all hydrogens were added, Gasteiger charges were computed and pdbqt files were created for docking simulations studies. The prepared receptors and ligands were used for molecular docking simulations.
Virtual screeningAutoDockVina® has reported high accuracy in predicting binding free energies by setting the receptor rigid while appraising flexible ligands with a comparatively low standard error [10, 11]. The ligands were docked into the receptors using AutoDockVina® and the virtual screening was done locally on a Linux platform using a script and configuration files containing information on the prepared receptors and ligands. The binding affinities were calculated and reported as mean ± SEM. Docked poses were visualized with VMD 1.9.1. [12]. Best poses were selected based on ligand intactness (some simulation models yielded broken up ligands), presence of ligand inside the binding pocket of the protein and binding free energy values. Search attempts were exhaustive. Four replicate docking runs were done for each tested compound.
Results and DiscussionBioinformatics mining of the PDB database afforded 49 structure hits (accessed 30th Jan 2015). The Zaire, Sudan and Reston species accounted for 30, 4 and 3 of these hits respectively. Five important Ebola virus proteins namely; VP24, VP30, VP35, VP40 and NP, were identified from literatures and their PDB codes 4bq0 (NP), moq (VP24), 2i8b (VP30), 4igb (VP35) and 1es6 (VP40) selected for further investigations. The results of the binding affinities of frontrunner antiviral compounds against Ebola proteins are presented in Table 2. In order to determine the frontrunner compounds, we used the binding affinities of favipiravir, (approximately 7.0 kcal/mol) to the Ebola protein targets as cut off value. Out of the 173 tested compounds, eleven compounds were found to bind strongly and concurrently on the five selected Ebola virus proteins. Eight compounds were selected for further investigations based on their binding affinities to Ebola viral protein targets. The ability of a compound to bind concurrently to more than one drug target is a great advantage because of the expected synergistic effect of such multi-target inhibitors. It is true that binding does not correlate with inactivation of function. Binding affinity is, however, a measure of the tightness with which a drug binds to the receptor. Binding alone does not determine the overall potency of a drug. It is one of the factors that influence potency. The other factor is efficacy. Efficacy does not fall within the context of this study. Studies on efficacy will be pursued in followup studies.
Ten plants featured prominently as possible sources of anti-Ebola phyto-constituents. We noted that three of the frontrunner compounds namely limonin, nomilin and hesperidin are compounds found in citrus species. Ursolic acid can be extracted from either Geum japonicum or Crataegus pinatifida. This means that ten plants (namely Hypericum perforatum, Stephania cepharantha, Phyllanthus amarus, Garcinia multiflora, Camellia sinensis, Citrus spp., Myrceugenia euosma, Rhus javanica, Geum ja ponicum, Crataegus pinatifida) could provide all the required phyto-constituents.
VP30 is a nucleocapsid-associated Ebola virus-specific transcription factor needed for transcription of the highly pathogenic Ebola virus [13]. A potential binding pocket for small molecules inhibitors is revealed in the structure [13]. Binding affinity energy values ranging between - 6.44±0.35 and -7.90±0.00 were obtained from the molecular docking simulations for VP30 against eleven (11) phyto-compounds.
VP35, an intrinsically disordered peptide of Ebola, has a great deal of potential as a new target for Ebola treatments, according to researchers at Washington University School of Medicine [14]. Inhibition of VP 35 leads to disruption in the emergence of newly created viruses from infected cells. Consequently, the virus did not survive.
There was no need for a no-Ebola protein as a negative control. This kind of negative control in molecular docking simulations is unnecessary, since a predefined search space must be specified for every protein drug target used for the simulation. This is not technically feasible when the protein target is absent.
The identified compounds represent significant lead to further investigation into the anti-Ebola activities of natural products. It is our considered proposal to promote the use of more than one of these natural products at the same time because of the expected synergistic effects.
There were previous reports of the use of Garcinia kola (bitter kola) as an anti-Ebola remedy. The garcinia kola compound was shown to halt multiplication of the virus in the laboratory. If this result is repeated in humans, the body would have a chance to fight off the virus. The discovery was announced at the 16th International Botanical Congress in St Louis in the US. According to the lead researcher, Prof M. Iwu, “the active compound is what is known as a dimeric flavonoid, which is two flavonoid molecules fused together.
Flavonoids are non-toxic and can be found in orange and lemon rinds as well as the colourings of other plants” [15]. Was it a coincidence that we found a garcinia biflavonoid (Robustaflavone) showing strong binding affinities for Ebola viral targets? Robustaflavone can be isolated form Garcinia multiflora. It is also not surprising that most of the identified phyto-compounds such as limonin, nomilin and hesperidin are flavonoids. These can be extracted from citrus species.
No virological data was generated in this study. The study is purely a computational one, otherwise known as Computer Aided Drug Design (CADD). Virological data will be generated in a follow-up study when we hope to gain access to a Bio safety Class 4 laboratory. Bio safety Class 4 laboratory is not available in my country presently.
Cost effectiveness is an important consideration in selecting sustainable health solutions. By using our natural resources, we may drive down the cost of treatment for the dreaded EVD. Reduced cost means that more persons will benefit from health care. This will ultimately free resources for other critical areas of growth and development. Use of natural resources is one way to break the disease-poverty cycle.
ConclusionOur study demonstrates that certain antiviral compounds found in several natural products may have potential in the management of EVD. Further investigations are needed to confirm these activities in relevant models. Are we close to solving the Ebola mystery? Only time will tell.
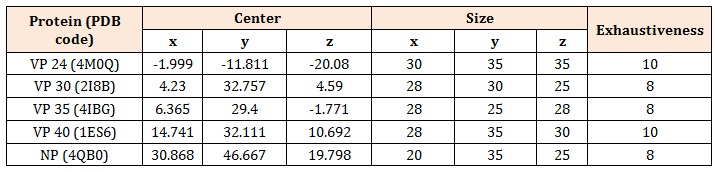
Table 1: Grid box centers and sizes of the five Ebola virus proteins.
Key: NP: NucleoProtein; VP24: Viral Protein 24; VP30: Viral Protein 30; VP35: Viral Protein 35; VP40: Viral Protein 40.
Table 2: Binding affinities of frontrunner antiviral compounds against Ebola proteins.
Key: NP: NucleoProtein; VP24: Viral Protein 24; VP30: Viral Protein 30; VP35: Viral Protein 35; VP40: Viral Protein 40
Chat with us on WhatsApp